Overview¶
General information and scope¶
The James Webb Space Telescope (JWST), a joint project by NASA, ESA, and CSA, is the successor mission to the Hubble Space Telescope. One of the four science instruments on board is the near infrared spectrograph NIRSpec. The NIRSpec Instrument Pipeline Software is a Python package that can be used for spectral extraction, background subtraction and radiometric calibration of both NIRSpec data and simulations.
The first development of NIPS (NIPS1.x) took place between 2008 and 2012 by Bernard Dorner and Pierre Ferruit. The software has been extensively used for nearly a decade for spectral extraction of data acquired during ground-test campaigns and simulations. In 2017, the decision to upgrade NIPS was made. This ensured the software’s maintainability (conversion to Python3, upgrade to use new external packages, significant code refactoring), but also equipped NIPS with a much wider range of capabilities, namely, background subtraction, treatment of individual targets, and all necessary steps to perform radiometric calibration in conjunction with a reference file repository. This development was carried out in 2018 by the core NIPS2.0 development team: Catarina Alves de Oliveira, Nora Luetzgendorf, Pierre Feirrut and Tim Rawle. The following pages will describe the implementation, installation, and usage of NIPS2.0.
Technical implementation¶
NIPS is a Python package comprising a module for the main pipeline class Pipeline, and additional modules for each pipeline step and its corresponding reference file and data classes. The graphic below depicts the overall structure of the python package and basic interplay between the modules.
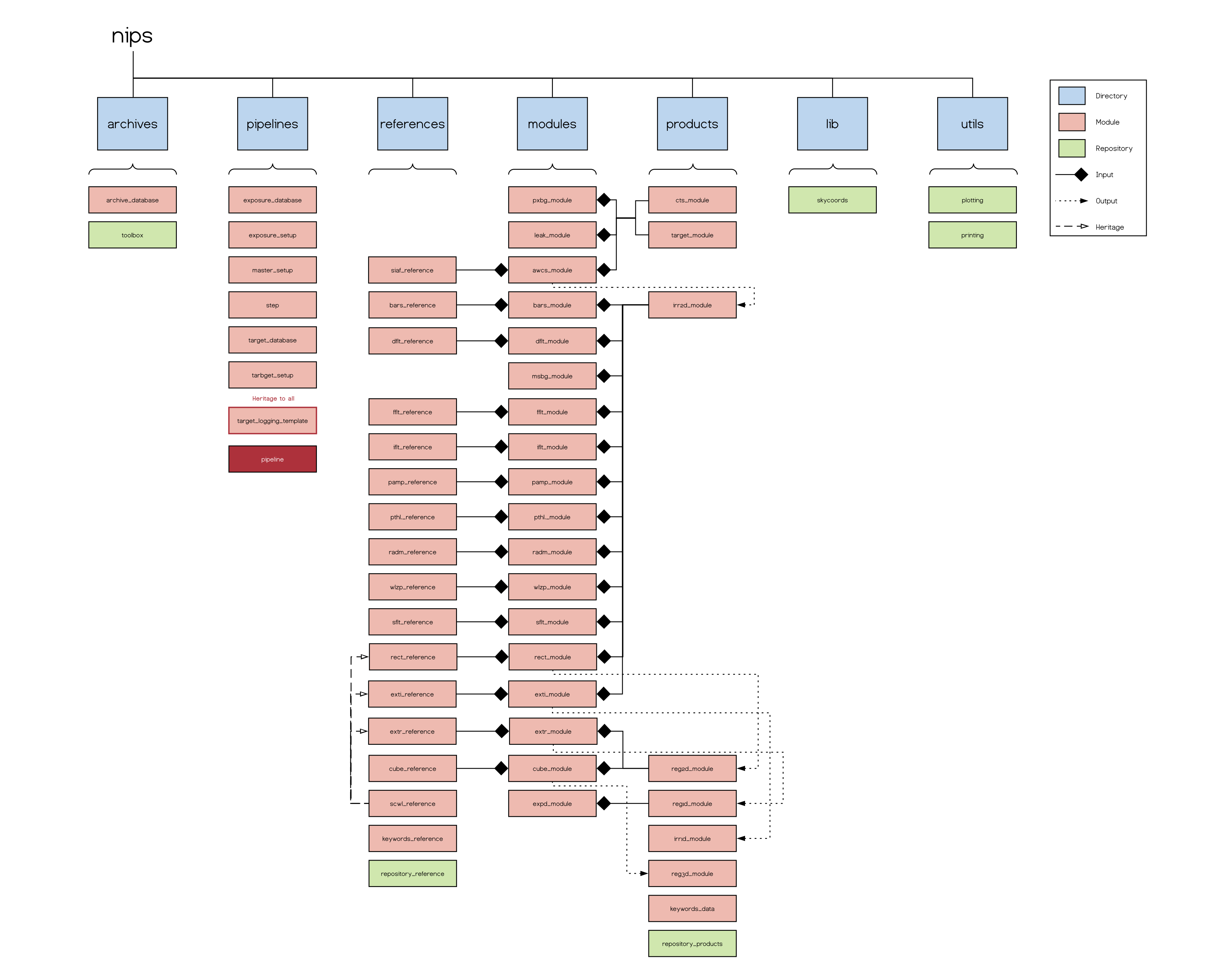
Although it is distributed as a separate package, it requires several other python modules and packages. For simplicity, NIPS is installed in a separate Anaconda Environment which avoids clashes with already existing python installations. To read more about Anaconda and how to install it please refer to installation section of NIPS (Installation and Setup) or the Anaconda documentation page.
Data handling¶
Data Archive and Database
Before the pipeline can run, an archive database has to be created for NIPS to be able to find the exposures based on NID and environments. NIPS then reads in the exposure file header, and constructs a corresponding instrument model from the reference file repository. The first step to run for each process is the awcs_module. This step extracts the traces of the desired aperture and assigns wavelength and y positions to each pixel in the trace. This is the basis of every following step.
Reference file repository
NIPS requires a standard repository for the calibration reference files used within the pipeline procedures. Further, it is intended that this standard repository will also be the primary location for storing all versions (including the latest) of all reference files, and the current consolidation project offers the perfect opportunity to standardize the organization and naming of the reference files themselves. All reference files are stored in the reference file repository. The repository ist located at the SVN and a local copy can be found on jpro and (name of computer at ESTEC?). More about reference files and their nomenclature can be found on livelink (ESA-JWST-SCI-NRS-DD-2017-001). The pipeline will always search for the best suited reference file based on the input data.
Targets
Targets (Target) are a new concept introduced in NIPS2.0. With launch approaching it is more useful to concentrate on an actual observation with a science target rather than an aperture. A target instance is created based on the observational parameters (most likely achieved from ATP). It holds information such as the type of source (point source, extended) and its location inside the aperture. For internal exposures the target will default back to the mode and slit names.
Steps
For the sake of more flexibility and structure, NIPS2.0 is based on a modular approach. Each pipeline step, dataproduct or reference file instance is handled in a different module. Like building blocks, those modules can then be used to dynamically set up a calibration flow dependent on the input parameters.
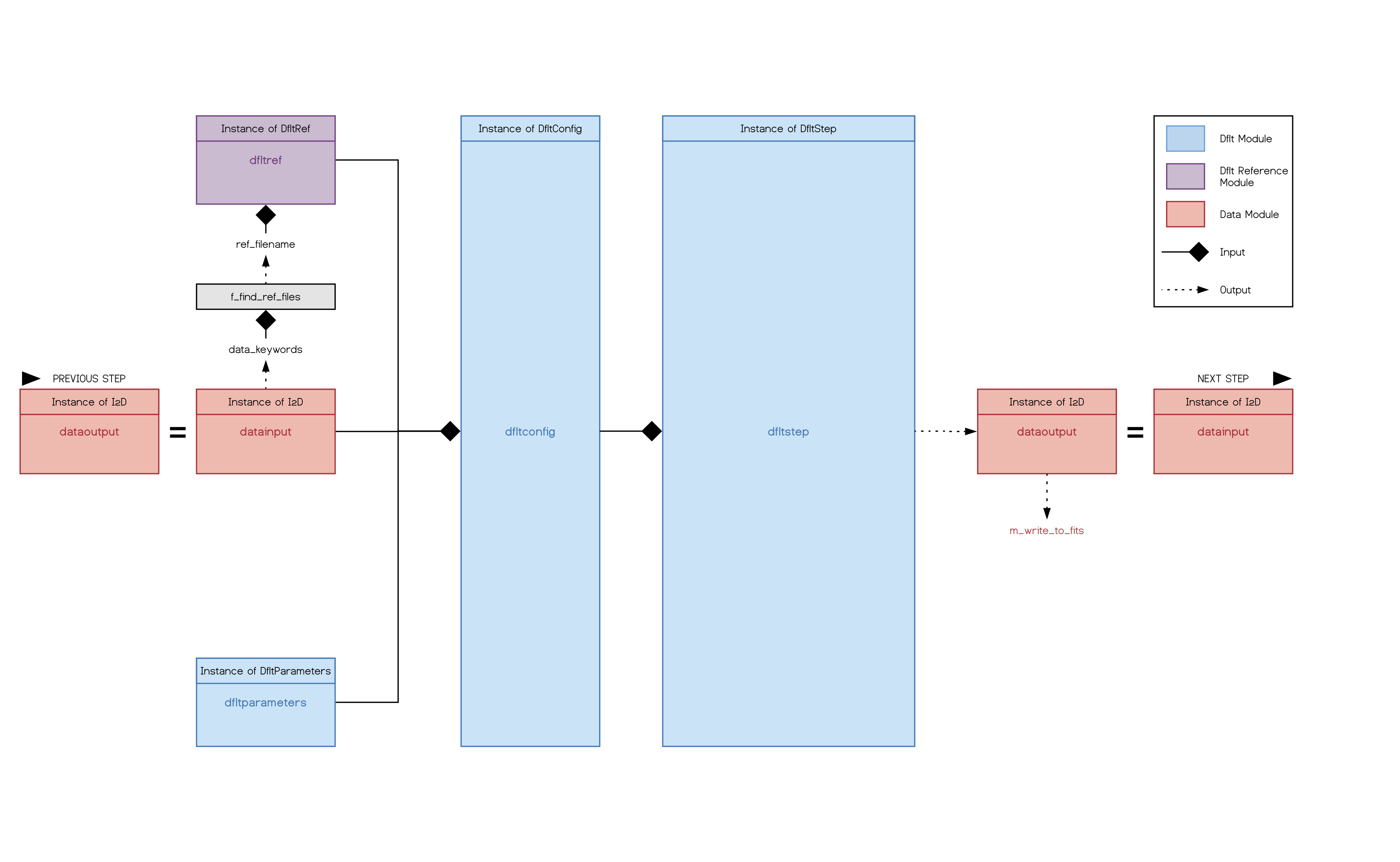
Each pipeline step follows the same schemata as displayed int he graphic below (for the D-flat step as an example). The input is taken from the previous step and the output given to the next step. The function f_find_ref_files() from the repository_reference will use the keywords from the input data to find the corresponding reference file in the reference file repository. This is passed to the reference module which handles the reading and writing of the reference files for each step. Reference class, data class and a parameters class (which stores the default values for each step, but allows modification, writing and reading of user specific parameters) are then parsed to the step module, where the actual calibration is processed on the input data. The pipeline step creates the output data and an extension of the applied calibration and parses it further down the pipeline. Depending of the output writing settings, this intermediate product can also be written out as a fits file.
Data products
The pipeline has six different data classes. One count-rate map class and five product classes. Those classes are independent of the observing mode (i.e. imaging, spectroscopy) but rather represent data types of different dimensions (1D, 2D, 3D) and if they are rectified or not (R, I). The name of a datatype/class is therefore composed from those two attributes. For example I2D is the data class for irregular 2D data (before rectification).
All output files are written in the configured output directory within a folder with the exposure ID. The structure is outlined below.
Note
When processing the same target with different parameters, a different target name has to be set, otherwise the existing data will be overwritten.
<output path>/
|-- config/
|-- config.json
|-- log/
|-- master_pipeline.log
|-- data/
|-- <NID>_<ENVIRONMENT>_<FWA>_<GWA>_<LAMP>/
|-- <NID>_<ENVIRONMENT>_<FWA>_<GWA>_<LAMP>_CTS1.fits
|-- <NID>_<ENVIRONMENT>_<FWA>_<GWA>_<LAMP>_CTS2.fits
|-- <NID>_<ENVIRONMENT>_<FWA>_<GWA>_<LAMP>.sqlite
|-- <NID>_<ENVIRONMENT>_<TARGETNAME>/
|-- config/
|-- pipeline_cfg.json
|-- target_cfg.json
|-- <step1>_cfg.json
|-- <step2>_cfg.json
|-- ...
|-- log/
|-- <NID>_<ENVIRONMENT>_<TARGETNAME>.log
|-- products/
|-- <NID>_<ENVIRONMENT>_<TARGETNAME>_AWCS_I2D.fits
|-- <NID>_<ENVIRONMENT>_<TARGETNAME>_RADM_I2D.fits
|-- <NID>_<ENVIRONMENT>_<TARGETNAME>_RECT_R2D.fits
|-- <NID>_<ENVIRONMENT>_<TARGETNAME>_EXTR_R1D.fits
In the main folder (called “master” by default), there is the master_pipeline log file which logs all main activities of the pipeline. For each target, there is a specific log file in the target folder as well as a collection of all parameters for each step, pipeline and target stored as json files. The pipeline accepts those json files as input if one wants to run the pipeline with the exact same configurations again. A more detailed description the naming and structure of the NIPS2.0 pipeline products can be found on livelink (ESA-JWST-SCI-NRS-DD-2017-002).
By default the pipeline outputs the 2D irregular product of the AWCS step (just after extracting and assigning coordinates), the last I2D product before rectification (if using the default pipeline steps, this will be after the RADM step), the rectified R2D product and the extracted R1D products or R3D cube in case of IFU. The PipelineParameters class accepts a keyword called PipelineParameters.writeout to control the output parameters.
Further reading¶
For more information about the specific calibration steps and their reference files we refer to ESA-JWST-SCI-NRS-TN-2016-008 and the references therein.